Data leakage
data leakage는 overly optimistic predictive model을 만들 수 있다. Training dataset밖에서부터 들어온 데이터로 인해 model이 결국 valid하지 못하게 develop되는 경우이다.
“if any other feature whose value would not actually be available in practice at the time you’d want to use the model to make a prediction, is a feature that can introduce leakage to your model”
“when the data you are using to train a machine learning algorithm happens to have the information you are trying to predict”
더 크고 복잡한 dataset일수록 data leakage문제가 더 심각해질 수 있다.
how to minimize data leakage when building a predictive mode:
perform data preparation within your cross validation folds
hold back a validation dataset for final sanity check of your developed models
(일반적으로, 위 두가지 technique들을 함께 사용해서 더 좋은 결과를 얻을 수 있다)
Preprocess data within cross validation folds
data전처리 과정에서 data leakage를 발생시킬 수 있다.
data normalization, standardization,등을 통해 전처리를 하고 cross validation을 통해 model의 성능을 확인하려한다면, data leakage로 인해 잘못 develop된 결과를 받을 수 있다.
The data rescaling process that you performed had knowledge of the full distribution of data in the training dataset when calculating the scaling factors (like min and max or mean and standard deviation). This knowledge was stamped into the rescaled values and exploited by all algorithms in your cross validation test harness.
non-leaky evaluation을 진행하기위해서는 rescaling을 위한 parameter calculation을 each fold of the cross validation에 각각 따로 진행할 수 있다. use those parameters to prepare the data on the held out test old on each cycle.
R의 경우 caret package와 scikit-learn의 경우 Pipeline을 통해서 re-preparing or re-calculating any required data preparation within your cross validation folds가 가능하다. feature selection, outlier removal, encoding, feature scaling and projection methods for dimensionality reduction, 등등이 cross validation folds내에서 진행되는 것이다.
Hold Back a validation dataset
더 심플한 방법은 dataset을 training과 validation sets로 나누어서 validation만 따로 hold back하는것이다.
Training set을 통해 modeling process가 모두 진행 된 이후 develop된 final model을 따로 둔 validation set을 통해서 평가하는 것이다. This can give you a sanity check to see if your estimation of performance has been overly optimistic and has leaked.
5 Tips to Combat Data Leakage
- Temporal Cutoff. Remove all data just prior to the event of interest, focusing on the time you learned about a fact or observation rather than the time the observation occurred.
- Add Noise. Add random noise to input data to try and smooth out the effects of possibly leaking variables.
- Remove Leaky Variables. Evaluate simple rule based models line OneR using variables like account numbers and IDs and the like to see if these variables are leaky, and if so, remove them. If you suspect a variable is leaky, consider removing it.
- Use Pipelines. Heavily use pipeline architectures that allow a sequence of data preparation steps to be performed within cross validation folds, such as the caret package in R and Pipelines in scikit-learn.
- Use a Holdout Dataset. Hold back an unseen validation dataset as a final sanity check of your model before you use it.
pipeline을 사용하는 예시
# import necessary library
from sklearn.pipeline import Pipeline
# declare the steps in our pipeline
pipe = Pipeline(steps = [('standardscaler', StandardScaler()),
('logisticregression',LogisticRegression())])
# fit the pipeline to our training data
pipe.fit(X_train, y_train)
# cross validate using our pipeline
cross_validate(pipe, X_train, y_train)
used a pipeline in conjunction with cross validation to ensure that as we cross validate, each fold of our cross validation is actually being scaled only on the data within the fold, then compared to the rest of the data, which is also scaled only on the data within their folds. This solves our problem of data leakage since we are not scaling our data across the entire dataset, then cross validating. Instead, we scale only on each fold, and cross validate on data also scaled within each of their folds.
# instead of using scikit learn's pipeline, we import from imblearn
from imblearn.pipeline import Pipeline
from imblearn.over_sampling import SMOTE
# just like we did for StandardScaler, we instantiate SMOTE within the pipeline
pipe = Pipeline(steps = [('smote', SMOTE(random_state = 42)),
('standardscaler', StandardScaler()),
('logisticregression', LogisticRegression())])
pipe.fit(X_train, y_train)
# cross validation using intra-fold sampling
cross_validate(pipe, X_train, y_train)
Much in the same way that scaling across an entire dataset will create data leakage, oversampling data based on the entire dataset will definitely leak data that shouldn’t be accessed by the training folds.
SMOTE와 StandardScaler를 pipeline에 포함시켜서 scaling과 oversampling이 done within each fold and not across all folds.
k-fold cross validation & stratified k-fold cross validation
stratified sampling 컨셉을 cross validation에 적용해서 training & test sets가 각각 original dataset과 동일한 class portions을 가지도록 설정할 수 있다. (training and the test sets have same proportion of the target variable.)
(straified sampling : “Stratified sampling is a sampling technique where the samples are selected in the same proportion (by dividing the population into groups called ‘strata’ based on a characteristic) as they appear in the population.”)
set the ‘stratify’ argument of ‘train_test_split’ to the characteristic of interest (e.g., target or class label variable)
K-fold cross validation
K-fold cross-validation splits the data into ‘k’ portions. In each of ‘k’ iterations, one portion is used as the test set, while the remaining portions are used for training.
Cross-validation implemented using stratified sampling ensures that the proportion of the feature of interest is the same across the original data, training set and the test set. This ensures that no value is over/under-represented in the training and test sets, which gives a more accurate estimate of performance/error.
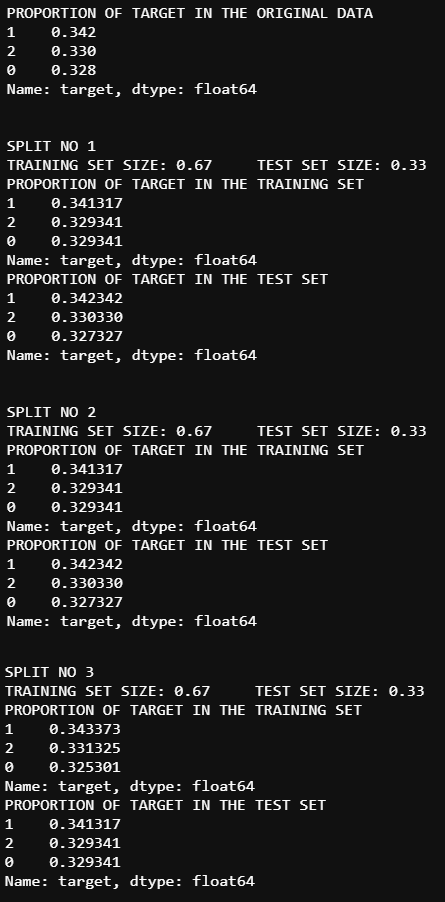
example code:
kfold = StratifiedKFold(n_splits=3,shuffle=True,random_state=11)
#data['target'] IS THE VARIABLE USED FOR STRATIFIED SAMPLING.
splits = kfold.split(data,data['target'])
print(f'PROPORTION OF TARGET IN THE ORIGINAL DATA\n{data["target"].value_counts() / len(data)}\n\n')
for n,(train_index,test_index) in enumerate(splits):
print(f'SPLIT NO {n+1}\nTRAINING SET SIZE: {np.round(len(train_index) /(len(train_index)+len(test_index)),2)}'+
f'\tTEST SET SIZE: {np.round(len(test_index) / (len(train_index)+len(test_index)),2)}\nPROPORTION OF TARGET IN THE TRAINING SET\n'+
f'{data.iloc[test_index,3].value_counts() / len(data.iloc[test_index,3])}\nPROPORTION OF TARGET IN THE TEST SET\n'+
f'{data.iloc[train_index,3].value_counts() / len(data.iloc[train_index,3])}\n\n')
output:
References
- https://towardsdatascience.com/the-right-way-of-using-smote-with-cross-validation-92a8d09d00c7
- https://vch98.medium.com/preventing-data-leakage-standardscaler-and-smote-e7416c63259c
- https://towardsdatascience.com/the-right-way-of-using-smote-with-cross-validation-92a8d09d00c7